Systems Biology
Virtual Liver
Our liver is the central metabolic factory taking care of glucose, lipid and nitrogen homeostasis and safeguarding us from toxins that we may have ingested. The liver fulfills these and further essential functions by means of a complex network of regulatory interactions and biophysical processes spanning molecular, cellular and tissue levels up to the whole organ. An understanding of liver functions and the rational design of therapies for diseases like liver cirrhosis have so far been limited by the high degree of complexity but shall now be enabled through a collaborative systems biology approach. The Virtual Liver Project is a novel initiative to explore this complex network and to iteratively construct a more and more detailed and predictive, mathematical, multi-scale model of the liver, the virtual liver. In close collaboration with experimentalists, our group explores the self-organizing mechanisms by which cells (e.g. hepatocytes and endothelial cells) establish and maintain the liver-typic tissue architecture (hepatic sinusoids) of sheets of hepatocytes that have access to three separate fluid systems, (i) blood from the intestine, (ii) highly oxygenized blood, and (iii) bile. Our multi-scale models link intra-cellular processes (endocytosis, membrane polarization, transcytosis) and inter-cellular processes (adhesion, signaling, motility, fluid transport). To account for the different processes, we have to combine various specialized modeling techniques including partial differential equations, structured population models and cell based models. We start out by predicting, analyzing and comparing the behavior of a few interacting cells in our models and in experiments of our collaborators. Subsequently, many cells with previously defined properties will be studied up to the scale of the liver lobule.
Cooperations:
Endocytosis
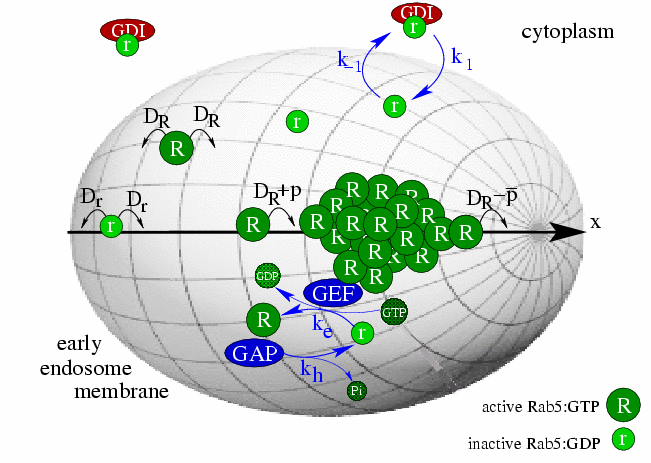
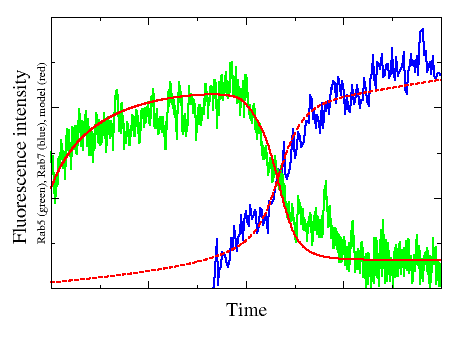
Endocytosis is a highly conserved process by which eukaryotic cells assimilate nutrients and process signals. Internalized material is transported by endosomes and sorted by means of endosome transitions. Biogenesis and maintenance of membrane identities as well as endosome transitions result from dynamic interactions among Rab GTPases. By applying a combination of techniques from bioinformatics, biophysics and mathematics, we try to discover the molecular regulatory mechanisms underlying transport in the endocytic pathway. Corresponding models are primarily based on (partial) differential equations and structured population equations.
Key Publications:
A general theoretical framework to infer endosomal network dynamics from quantitative image analysis
Curr. Biol.,
22,
15,
1381 - 1390,
2012
[DOI]
Membrane identity and GTPase cascades regulated by toggle and cut-out switches
Mol. Syst. Biol.,
4,
206,
2008
[DOI]
Cooperations:
Prof.
Frank Jülicher (Max Planck Institute for the Physics of Complex
Systems, Dresden)
Prof.
Marino Zerial (Max Planck Institute of Molecular Cell Biology and
Genetics, Dresden)
SpaceSys
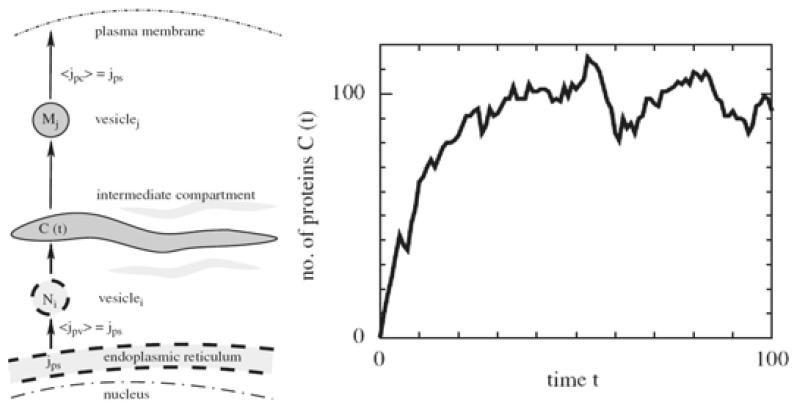
The aim of the junior research group "SpaceSys - A Spatio-temporal Approach to Systems Biology" is to further build on live cell imaging data together with mathematical models and analysis to develop new biological insight at the interface between cell biology and developmental biology. SpaceSys will tailor methods for spatially dependent processes such as partial differential equation models, integro-differential equations and bifurcation theory known from applied mathematics and physics to be applicable in Systems Biology. Specifically, we address questions from sub-cellular compartmentalization and secretory transport to questions of morphogen patterning and tissue size regulation.
Key Publications:
Ligand-Specific c-Fos Expression Emerges from the Spatiotemporal Control of ErbB Network Dynamics
Cell,
141,
884-896,
2010
[DOI]
Cooperations:
Prof.
Thomas Höfer (German Cancer Research Center, Heidelberg)
Prof.
Udo Reichl (Max Planck Institute for Dynamics of Complex Technical
Systems, Magdeburg)