This is an old revision of the document!
Cellular Potts models
Differential Adhesion: cell sorting in two dimensions
Introduction
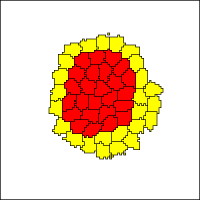
This model shows the original cellular Potts model (a.k.a. Glazier-Graner model) of cell sorting based on the Steinberg's differential adhesion hypothesis.
Model description
Two CellTypes are defined, each of which has a VolumeConstraint specifying the cell's target area/volume. In the CPM element, the MetropolisKinetics can be configured and the Interaction energies between cell types are specified.
Although cells can be initialized as single points using e.g. the InitCircle plugin, in this example, the Nodes of each Cell in the CellPopulations are given explicitly. In fact, these Populations are restored results of a previous simulation.
The simulation shows two populations of spatially resolved cells that initially organized in a mosaic fashion. Through differential adhesion, the motile cells sort out and re-organize into an distribution in which one cell type engulfes the other.
Snapshots of the simulation are saved to files named [Title][Time].xml.gz. These files containing intermediate and result states can be opened and used as initial conditions for new simulations. Remember to change StartTime and StopTime accordingly.
Model
h CellSorting_2D.xml |h
extern>http://imc.zih.tu-dresden.de/morpheus/examples/CPM/CellSorting_2D.xml
Examples → CPM → CellSorting_2D.xml
Reference
Graner F, Glazier J. Simulation of Biological Cell Sorting Using a Two-Dimensional Extended Potts Model. Phys. Rev. Lett. 69 (13): 2013–2016, 1992.
Proliferation in two dimensions
Introduction
This model show a simulation of a growing cell population, using the cellular Potts model.
Model description
This model specifies CellType which has a VolumeConstraint and a Proliferation plugin.
In the Proliferation plugin, the Conditions for a cell to divide are given. Here, each cell that has more than 90% of the target volume has a small probability to divide. Once a division has taken place, the Equation defined in the Triggers elements are triggered.
Model
h Proliferation_2D.xml |h
extern>http://imc.zih.tu-dresden.de/morpheus/examples/CPM/Proliferation_2D.xml
In Morpheus GUI: Examples → CPM → Proliferation_2D.xml
Persistence
Introduction
This example shows self-organized collective motion of cells as a result of persistence ('cellular inertia'). A similar model has recently been used in (Czirok et al., 2013).
Description
The model uses the Persistence plugin that causes cells to prefer to move in their current direction. The direction is stored in a PropertyVector that is used to plot the color and arrows in Gnuplotter.
The model is simulated in a circular domain with constant boundary conditinos, which can be set up in Lattice / Domain / Circle. The value for the constant boundary is specified in CPM / BoundaryValue.
Try it!
- Change the boundary conditions from circular=constant to x/y=periodic and observe the resulting collective motion.
- Change the
decay-timeofPersistence(specifying the 'memory').
Model
h Persistence_2D.xml |h
extern>http://imc.zih.tu-dresden.de/morpheus/examples/CPM/Persistence_2D.xml
Examples → CPM → Persistence_2D.xml
Reference
Czirók A, Varga K, Mehes E, Szabo A, Collective cell streams in epithelial monolayers depend on cell adhesion. New J. Phys. 15 075006, 2013.
Proliferation in three dimensions
Introduction
This model show a CPM simulation of a growing cell population in 3D.
Model description
This model specifies CellType which has a VolumeConstraint and a Proliferation plugin. In the Proliferation plugin, the Conditions for a cell to divide are given. Here, each cell that has more than 90% of the target volume has a small probability to divide. Once a division has taken place, the Equation defined in the Triggers elements are triggered.
In this model, two medium cell types have been defined. One of these (called matrix) is used to represent a matrix with higher adhesivity. This is done by (1) defining the 'matrix' cell type as a BoundaryCondition of the -z boundary in the CPM and (2) providing lower contact energy for cell-matrix interaction than for cell-medium interactions.
The simulation is visualized using the TiffPlotter that saves TIFF image stacks that can be loaded by image analysis software such as Fiji and displayed using Fiji's 3D Viewer plugin.
Model
h Proliferation_3D.xml |h
extern>http://imc.zih.tu-dresden.de/morpheus/examples/CPM/Proliferation_3D.xml
In Morpheus GUI: Examples → CPM → Proliferation_3D.xml
Run and Tumble
Introduction
This example models a single cell that moves according to a Levy walk: a random walk with occassional occurrence of long straight walks.
Model description
The model defines a CPM cell that has two properties:
- A
PropertyVectorthat gives the direction of movement and - A
Propertythat defines the time when this direction of movement is changed.
The change in direction is using a VectorRule. In this case, it specifies a new random direction for each of the 3 x,y,z coordinates separately: move_dir = sin(angle), cos(angle), 0 where angle = rand_uni(0, 2*pi).
This is calculated with an Event. Upon triggering, this sets the new direction and a waiting time until the next change of direction. To model a superdiffusive Levy walk, this waiting time is chosen from an exponential distribution: change_time = time + 20 * rand_gamma(0.5, 5)
Finally, the cell is made to move in the chosen direction using DirectedMotion that takes the PropertyVector as input.
Model
h RunAndTumble.xml |h
extern>http://imc.zih.tu-dresden.de/morpheus/examples/CPM/RunAndTumble.xml
In Morpheus GUI: Examples → CPM → RunAndTumble.xml
Stem cells in the intestinal crypt
Introduction
This illustrative example shows the emergence of clonal populations from stem cells in an intestinal crypt. Stem cells in the bottom of the crypt divide asymmetrically and produce a population of transit amplifying (TA) cells. For each of the TA cells, the color indicates the stem cell is it derived from.
Model description
This model shows several new modeling features, available as of Morpheus 1.2.
Loading domain from image
The crypt-like domain is specified by loading an external 8-bit TIFF image file using Lattice/Domain/Image.
Asymmetric cell division
Stem cell divide asymmetrically using the new ChildID handles in the Proliferation plugin. This sets a user-defined symbol (here: daughter) to either 1 or 2 that can be used to distinguish both daughter cells. This symbol is then used to set the stemness s of one daughter to 1 and the other to 0: s = if(daughter==1, 1, 0).
Conditionally changing cell types
When a cell looses its stemness s, it is moved to the TA cell type. This is done using the new ChangeCellType plugin.
Upon satisfyiug its Condition, ChangeCellType move the cell to te specified new cell type. By default, all the properties of a cell that exist in both cell type context are maintained and nuspecified ones are set to their default values. This default behavior can be overridden in Triggers by specifying Rules stating how to deal with specific properties.
PopulationReporter
The new PopulationReporter allows the collection of statistical data about the cell population. Here, it is used to count the sizes of the various clonal populations. This number of reported into a Global and subsequently written to file and plotted using a Logger.
Model
h Crypt.xml |h
extern>http://imc.zih.tu-dresden.de/morpheus/examples/CPM/Crypt.xml
In Morpheus GUI: Examples → CPM → Crypt.xml